-
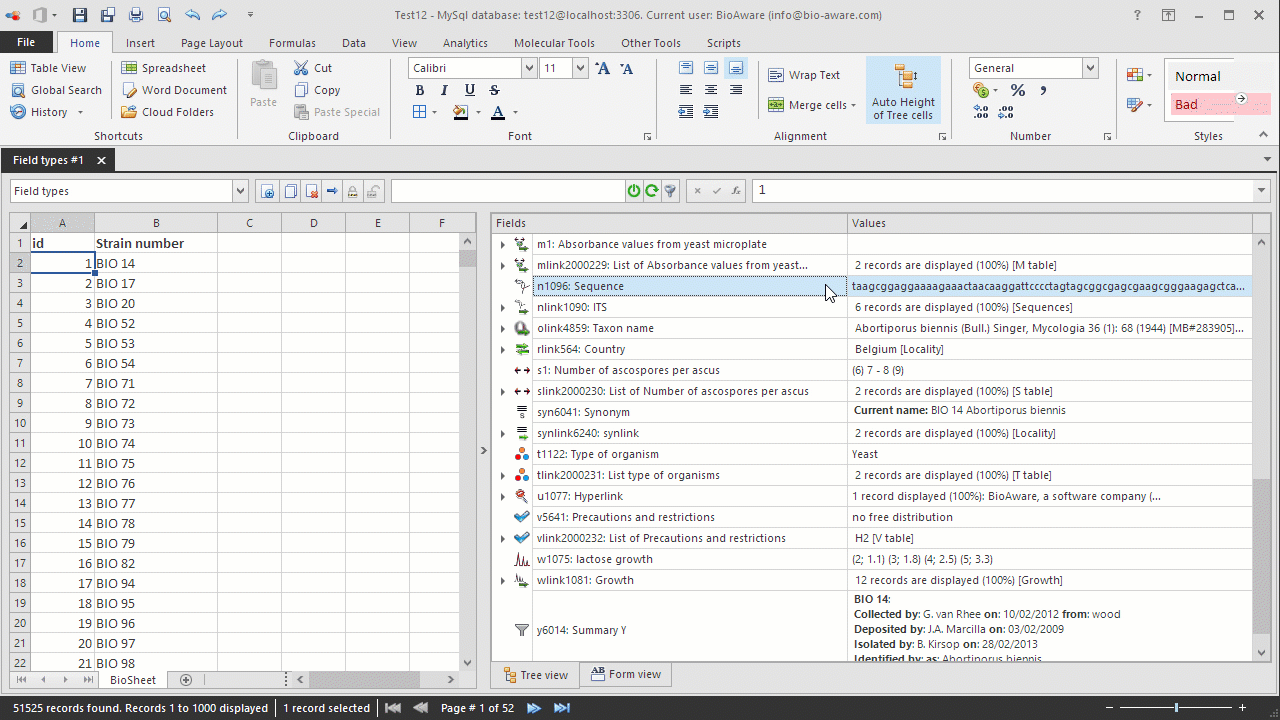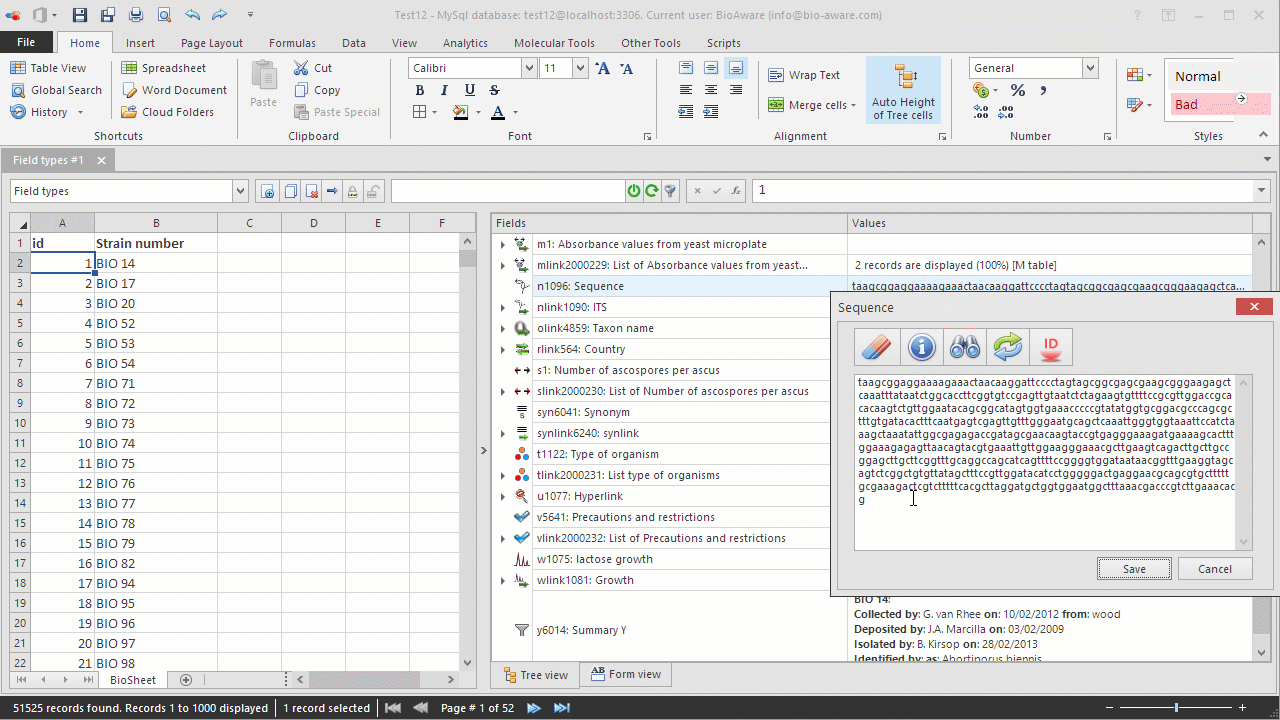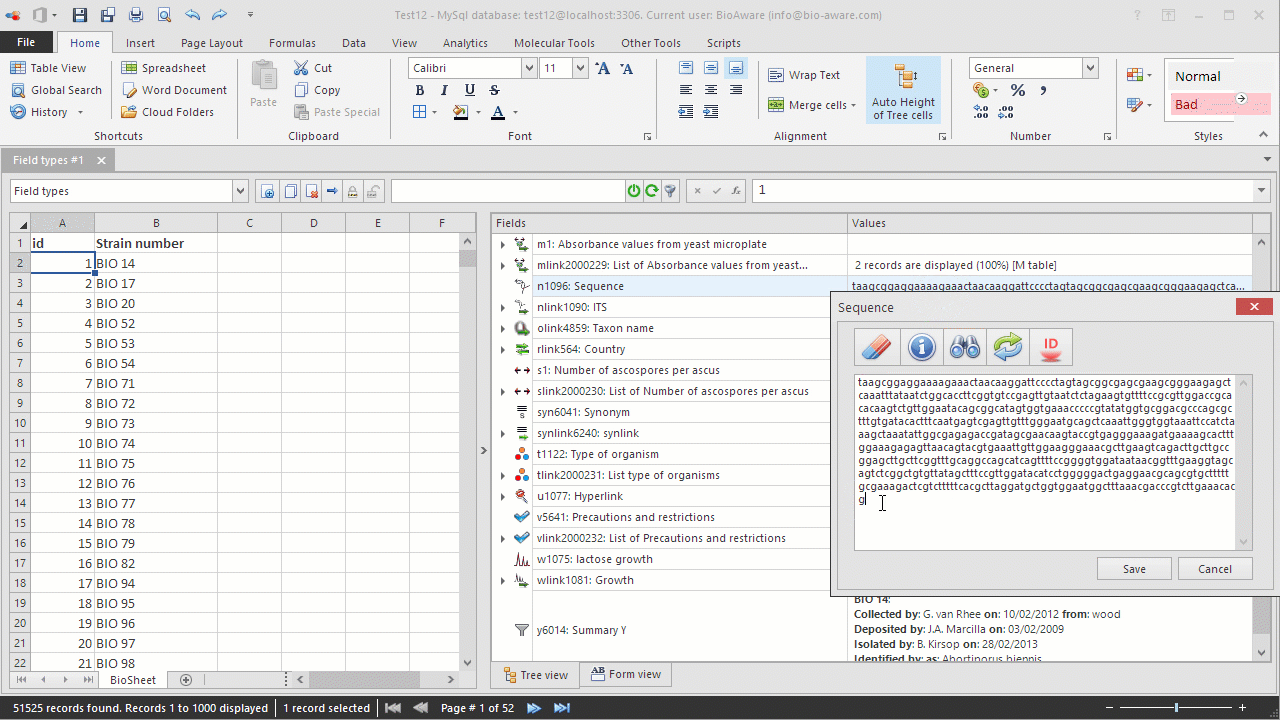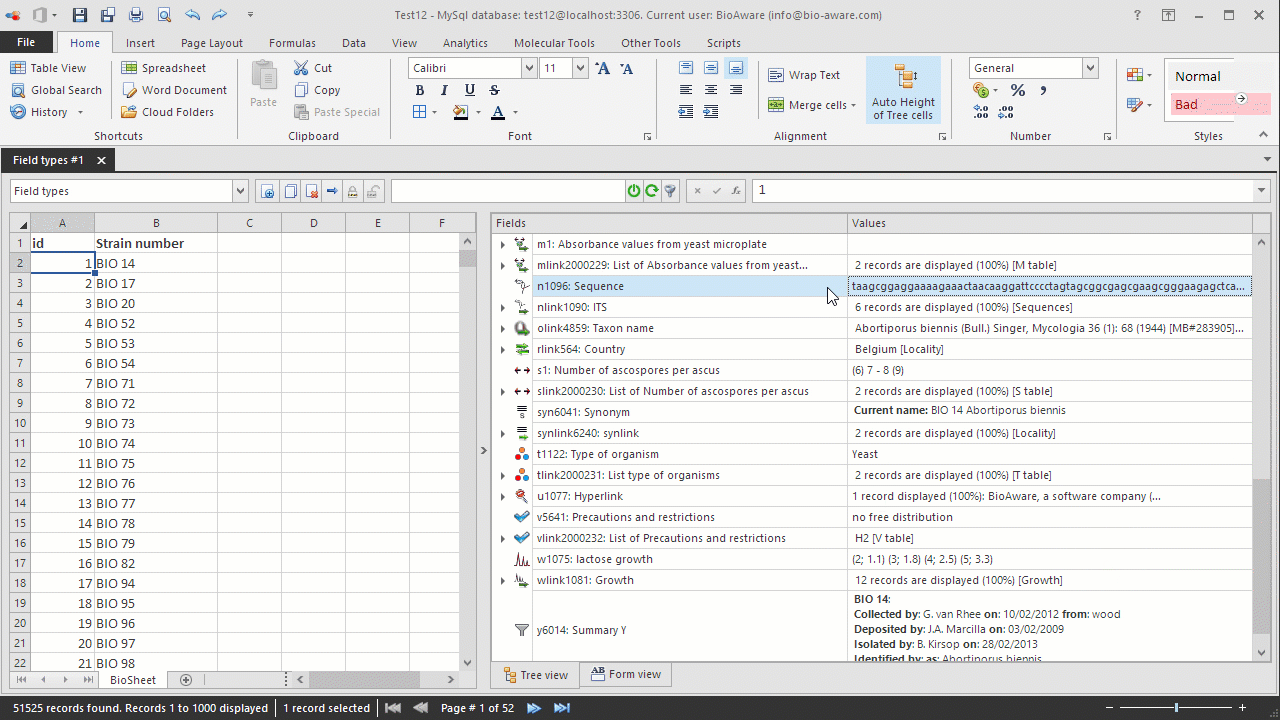
Select the record(s) to be edited in the BioSheet on the left side of the window.
-
Select the N field to be edited in the tree on the right side of the window.
-
Double-click on the field's Values column. A popup window will appear.
-
Paste the sequence in the textbox.
-
Additional function are available from the popup:
-
Click Save to save the sequence.
Note that several records (multiple selection) can be edited at the same time for a given field. Also, several fields having the exact same properties can be edited at the same time using the multiple field selection feature.
Click Ctrl + Z to undo the last change; Click Ctrl + Y for redo.
 Clear
sequenceTo remove the unwanted characters from a given sequence such as numbers, carriage-return or letters that cannot be present in a sequence.
|
|
 Statistics To obtain information related to a sequence such as the length, the number of A, C, T, G or the possible conversions into the protein (for DNA sequences only).
|
|
 SearchSearch DNA segment in this sequence.
In the popup, enter the substring to search for in the full sequence:
The results will be shown including
-
The number of time the substring was found. -
Sequence length of the full sequence in this field for the selected record. -
The exact position(s) where the substring was found. -
And it highlights the found substring with red font.
|
|
 Reverse-complement Reverse-complement this sequence.
To reverse-complement a sequence means to perform two operations on a DNA sequence:
-
Reverse the sequence — flip it from 5′→3′ to 3′→5′.
-
Complement it — replace each base with its complementary base (A ↔ T and G ↔ C).
For example:
Input: 5' - -A C T G A A -3'
Reverse: A A G T C A
Complement: T T C A G T
Reverse complement: 5'- T T C A G T -3'
|
|
 Identification
with BlastTo compare a sequence with another one or to identify it against an existing database. For more details, see Identification.
|
|
 Select entire
sequenceSelect entire sequence, even if too big to be displayed.
|
|
 Select
genetic code
Translated DNA to proteins using one of these genetic codes:
Choose from: Standard, Mitochondrial, Yeast Mitochondrial, Invertebrate, Ciliate Nuclear, Echinoderm Mitochondrial, Mycoplasma, Bacterial.
DNA sequences are being translated into protein sequences (amino acids) using different genetic codes, depending on the organism or cellular context.
Background information:
-
Translation is the process of converting a DNA (or mRNA) sequence into a protein. -
This is done using the genetic code, which defines how triplets of nucleotides (codons) correspond to amino acids. -
While the Standard Genetic Code is universal for most organisms, some organisms use slightly different codes (called variant genetic codes).
What does each option refers to:
|
Genetic Code Option
|
Used By
|
Differences?
|
|
Standard
|
Most organisms (nuclear DNA)
|
Default code
|
|
Mitochondrial
|
Human and mammalian mitochondria
|
Some codons have different meanings (e.g., UGA = Trp)
|
|
Yeast Mitochondrial
|
Mitochondria in yeast species
|
Unique changes
|
|
Invertebrate
|
Mitochondria of invertebrate animals
|
Slight differences from human mito code
|
|
Ciliate Nuclear
|
Nuclear DNA of ciliates (e.g., Paramecium)
|
Stop codons may be reassigned
|
|
Echinoderm Mitochondrial
|
Sea stars, sea urchins, etc.
|
Specific mitochondrial code
|
|
Mycoplasma
|
Certain bacteria with small genomes
|
Alternative codon usage
|
|
Bacterial
|
Most bacteria (prokaryotes)
|
Similar to Standard, some variations
|
Why does this matter?
When translating a DNA sequence to a protein, you must use the correct genetic code. If you use the wrong code:
-
You may get wrong amino acids. -
You might translate a stop codon incorrectly. -
The resulting protein could be nonfunctional or misleading.
|
|